TL;DR Using functional information improves fine-mapping in genome-wide association studies
I recently reviewed a paper title “Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping Studies”, which has has now been published. Overall I thought this was a useful contribution that improves on several methodological aspects of fine-mapping in genome-wide association studies.
(NB: none of the points from my actual review are still worth discussing, so the following are my somewhat rambling current thoughts)
Key points
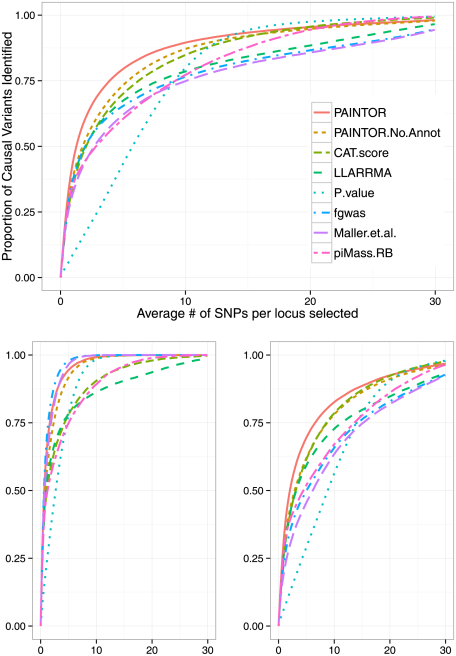
The key to the method here is that 1) it explicitly includes a prior from functional genomic information and 2) it allows for multiple causal SNPs. The figure at the top of this post shows how different methods perform in identifying truly causal SNPs in simulations–in the bottom left is performance in simulations with a single causal variant at a locus, and in the bottom right performance in simulations with multiple causal variants. Perhaps unsurprisingly, methods that explicitly assume a single causal variant (like fgwas, which I wrote) perform best in the former situation, while methods that allow multiple causal variants (like PAINTOR, by these authors) perform best in the latter situation.
Next steps
One thing that I’ve been thinking about is the question: how much do we expect to gain from incorporating functional genomic information into GWAS? In this study, the authors are able to reduce the number of plausible causal variants at a locus by around ~20% using annotations enriched around 5-10X for strong associations (see also a paper from Gusev et al. with similar results); in my own work, I’ve seen that this style of approach also increases the number of identified associations by around 5%. This shows that this line of work is on the right track, but personally, I’d perhaps naively expected this approach to be more powerful. A couple possibilities:
- Do we have the right functional annotations? One possibility is that we don’t have data from the tissues and experimental conditions that are most relevant to annotate disease-related SNPs–for example, perhaps we need to incorporate maps of transcription factor binding in stimulated immune cells, or from different developmental stages. Some nice work along these lines has been done by Fairfax et al.
- Do we need to get better at predicting which SNPs in an annotation are important? Another possibility is that we’re doing a poor job of distinguishing SNPs that matter from SNPs that don’t; e.g. two variants could both fall in an annotated transcription factor binding site, but only one might actually influence binding. Important work along these lines has been done by Moyerbrailean et al.
It seems unlikely that there’s going be be a “magic bullet” that works in all situations; rather, it will take progress on each of these points, plus the development of new methods that use all these different sources of information together.
Thank you for your review! I share your thoughts on fruitful future directions and let me also add several of my own thoughts to your list:
3. Existing methods make an explicit/implicit assumption that functional annotations have the same weight across all risk loci. This assumption might be invalid in some cases. For example, one could argue that existing GWAS loci are biased towards common variation whereas as new, to be identified, GWAS loci might be skewed towards low-frequency variation and one could envision functional annotations with different optimal weights in those cases.
4. Part of the lost signal comes from requiring the methods themselves to select the “correct” annotations. Improvements could be made by reducing the initial set of plausible annotations. Variance-components approaches such as Gusev et al seem the most promising in that direction.
5. As part of other works, we consistently see imputation increasing the functional enrichment signal so maybe with more variants accurately imputed entering the model we might see increased relative performance from functional annotations.
Best,
Bogdan
Hi Bogdan,
Thanks for the additional thoughts. The idea about estimating different enrichment parameters for variants at different frequencies is a particularly interesting one (i.e. maybe common nonsynonymous SNPs should be treated differently than rare nonsynonymous SNPs), definitely worth following up on.
Joe
Thank you for this post.
I agree that better functional analysis and imputation will be key to any GWAS-functional analysis. But I also think we will need to get better at modelling that functional annotation. Everyone has seen enrichment for cell-specific states, but those cellular states have a heirarchy: CD4s are much closer to CD8s than to megakaryocytes. Certain TFs are much more relevant to disease than others (eg, for T1D: IKZF3, BATF and ESRRA [1]), and their relevance probably varies in a state specific manner. A SNP in a particular regulatory region may only affect disease if that regulatory region hits a gene in a disease relevant pathway. This relates to Bogdan’s point 3, but I think it’s more complicated than SNP MAF. HiC data will help us assign target genes to regulatory sequences in the near future, but pathway knowledge is still woeful. Yet somehow, I think, we need to pull in all these strands of information in our model of functional information. These are themselves high dimensional, highly structure data, and we need to work out how to model this in order to properly integrate GWAS results.
1. http://bioinformatics.oxfordjournals.org/content/early/2014/09/18/bioinformatics.btu571.long