(Short answer: yes)
In my recent paper describing a hierarchical model for genome-wide association studies, I present estimates of the proportion of GWAS hits for different phenotypes that are driven by non-synonymous variants. A colleague recently wrote to tell me there was some debate in his journal club about whether these estimates (and other such estimates in the paper) are unbiased. I had simply assumed this was the case, so there is no discussion of this point in the paper.
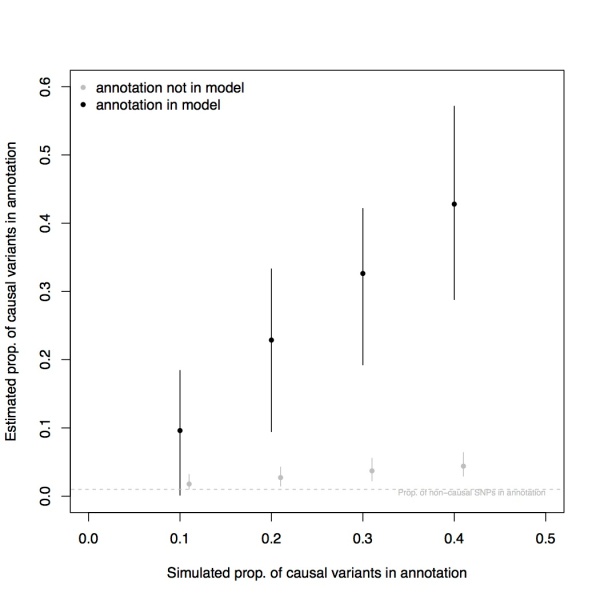
To test whether this intuition is correct, I performed simulations of an association study of a quantitative trait with ~100 causal variants, and I assigned causal variants to a simulated annotation as varying rates (Instead of a detailed description of the simulation methods, I’ve posted my code to GitHub). I then ran fgwas and estimated the proportion of causal variants in that annotation, and then repeated this simulation 100 times. Shown in the Figure above are the range of these estimated proportions in the 100 simulations, excluding the 10 most extreme estimates (the 5 highest estimates and the 5 lowest estimates).
The estimates do appear to be unbiased in these simulations (as well as simulations with a higher baseline rate of non-causal SNPs in the annotation, see here). By contrast, a naive estimator of the fraction of SNPs in the annotation that does not use knowledge about enrichment (in grey) is a severe underestimate.