tl;dr things cost money
It’s not news that most bioinformatics resources are short-lived. Websites mentioned in scientific papers go offline at about a rate of 5% per year, and even funding for extremely popular resources like the HapMap Browser can dry up effectively overnight. Indeed, researchers who work on model organisms are currently begging the National Institutes of Health to continue supporting basic informatics infrastructure.
Here, I want to discuss whether this is actually a problem (I’d say yes), one possible solution (paying for things), and one specific toy example of how this could work (a bitcoin-payable interface to the Exome Aggregation Consortium [ExAC] data).
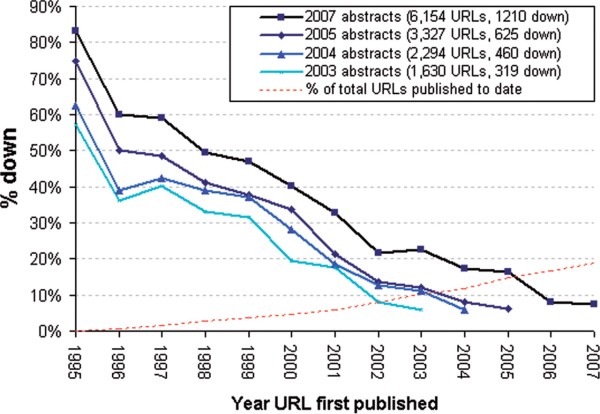
Link rot in the medical literature, from Wren 2008
Is there really a problem, and if so what is it?
There’s a legitimate argument to be made that there’s not really a problem. After all, most papers are rarely cited, so it stands to reason that most software packages or web services are rarely used. And if they’re not used, why should they be supported by grant dollars? Further, the most widely-used resources do in fact get non-trivial levels of funding (the model organism databases discussed above will still get millions of dollars in grants even if the proposed funding cuts go through). So maybe everything is hunky-dory.
However, what this misses is there are a number of important resources that are unlikely to win grants simply because they’re not “innovative” or “novel”, but rather the more mundane “useful”. There are few mechanisms available to sustain projects like this (as has been pointed out many times). More importantly, there is little incentive to actually undertake projects like this–I would probably recommend against an academically-minded student or postdoc taking on a project like “re-implement the HapMap Browser”.
So maybe the problem can be phrased as: we (the scientific community) want two somewhat contradictory things–we want grant money to go to innovation and scientific discovery, and at the same time we want people to maintain resources that work well just the way they are. Any attempt to balance those two things inevitably is going to lead to conflict, and in general I think it’s likely that the former will win out over the latter just on political grounds.
A potential solution: paying for things
Titus Brown alludes to this problem in his post Sustaining the development of research-focused software:
The lesson we can take from the open source world is that, in the absence of a business model, only “community project” software is sustainable in the long term.
The other obvious sustainability model here is of course right there–how about users start paying for things?
The “users paying for things they like” model could even have beneficial consequences beyond simple cash. When The Arabidopsis Information Resource [TAIR] started charging for access, director Eva Huala explained (as paraphrased in Science): “As a bonus…because TAIR doesn’t rely on federal grants, it no longer has to please peer reviewers and can focus instead on what users want”. This comment suggests that perhaps grants are a poor mechanism for funding these types of resources (assuming, of course, that the goal is to make things that users want!).
But charging for access to a database or software has some serious problems–you have to set up an entire billing and sales infrastructure, and even a modest paywall probably reduces your number of users (and thus the utility of your work to the scientific community) by orders of magnitude.
One suggested fix to these problems is “micropayments”–making paywalls so small that they’re not even worth thinking about. Historically this has proved difficult. As Nick Szabo has pointed out, the mental cost of simply thinking about whether something is worth paying for can become the limiting cost in a transaction. Further, interchange fees charged by credit card companies put a lower limit (of at least a few cents) to the size of micropayments that are feasible when using credit cards.
In this context I’ve been intrigued by the work done by the company 21 [1]. Instead of worrying about credit card systems and international wire transfers, they work with Bitcoin, the “money over IP” system. They’ve set up an infrastructure that allows micropayments of a little as a single satoshi (at exchange rates when I wrote this sentence, this is 0.0006 US cents), and software that allows users to trivially get set up with a bit of bitcoin to play with.
How could this type of technology help with the problem I mentioned before?
Toy example: a Bitcoin-payable API for the ExAC database
As a toy example, I threw together a simple bitcoin-payable API for the Exome Aggregation Consortium data. If you’ve installed the 21 software, getting a list of loss-of-function variants in a gene (along with their allele frequencies and a bit of additional information) is then as simple as, for example:
> 21 join
> 21 buy url http://10.244.188.146:5000/lof/PCSK9
Alternatively the API can be called and paid for using the 21 python library. Each API call costs 1000 satoshis (around half a cent), and I’ve implemented endpoints that pull out all annotated variants in a gene as well as all loss-of-function variants (again, this is just a toy example).
All of this is running on an Amazon EC2 micro instance, which will set you back something like $75/year. If the ExAC Browser gets a couple million page views a year, then charging 1/50th of a cent per page view (about 75 satoshis) could cover a few instances, or 1 cent per page view would net $20k, enough for basic electricity plus a bit to play with [2]. In principle, Amazon (or some future cloud computing provider) could set up automatic payments in a system like this, such that any resource that earns enough to cover costs might be maintained on long time scales without human intervention.
Of course, the real benefit here is that once bioinformatics resources start paying for themselves, you can start to build on top of them with a bit more assurance that they won’t disappear overnight. A reliable and self-sustaining infrastructure might open up exciting new possibilities.
—
[1] I have no financial relationship with this company. I do own some bitcoin though; by NIH standards I have a “significant financial interest” in the currency.
[2] I suspect there would be social pressure against anyone actually making a profit on something like this, but I personally wouldn’t have any major objections to students making beer money (I suspect this is the order of magnitude that is feasible in most cases) by building useful tools.